Sleep EEG Analysis
Modern sleep science is fundamentally informed by ExG biosensors recording brain activity (electroencephalography, EEG), eye-movement activity (electrooculography, EOG), and muscle activity (electromyography, EMG) to characterize the physiology of sleep.
Here, we demonstrate how EEG data recorded during a full night’s sleep using Mentalab Explore can be analysed with the free and open source yasa toolbox. We will perform automatic sleep staging by applying the yasa classifier [1] to EEG data, inspect band power in the EEG signal, and detect slow waves and sleep spindles. The application example follows the excellent yasa notebooks.
Data acquisition
Our volunteer recorded their sleep using an 8-channel Mentalab Explore device. The following montage was applied:
Channel Type Electrode Position
0 REF adhesive TP10 (right mastoid)
1 EOG adhesive above right eye
2 EOG adhesive outer canthus right eye
3 EMG adhesive submental (below chin)
4 EEG dry O2
5 EEG dry C2
6 EEG dry F2
7 EEG adhesive C1
8 EEG wet Fc1
Clip cables were used to connect to adhesive and dry electrodes. Dry electrodes use flexble brushes to establish connection. Impedances were not lowered after positioning the electrodes. Refer to our sleep recording protocol, for detailed reccomendations on preparation, data transfer, and on how to convert the recorded BIN file to .csv or .bdf files.
Sleep data analysis
Software preparation
First, we prepare a conda virtual Python environment to house our project.
- (Install anaconda)
- Create a virtual python environment using conda:
conda create -n myenv python=3.10 - Activate your conda environment:
conda activate myenv - Install the required packages:
pip install numpy mne yasa
Data processing
We then load the required packages in python.
import mne
import yasa
import numpy as np
import matplotlib.pyplot as plt
We will load a CSV file containing the sleep data recorded using Mentalab Explore and convert it to an mne raw data object. The code example expects data recorded at 250 Hz with an 8-channel device. It is important to note that mne expects data in Volt (V), whereas Mentalab Explore stores data in Microvolt (µV).
ch_names = ['EOG-above', 'EOG-canthus', 'EMG-chin', 'Dry-O2', 'Dry-C2', 'Dry-F2', 'Sticky-C1', 'Wet-Fc1']
data = np.loadtxt("./data/wiki-sleep-analysis-data/Mentalab-sleep-analysis_ExG.csv",skiprows=1,delimiter=',').transpose()[1:9]
sf = 250
ch_types = ["eog", "eog", "emg", "eeg", "eeg", "eeg", "eeg", "eeg"]
info = mne.create_info(ch_names = ch_names, sfreq = sf, ch_types = ch_types)
raw = mne.io.RawArray(data, info)
raw.apply_function(lambda x: x * 1e-6) # scale muV to V
Next, we apply a band-pass filter with a low cutoff of 0.1 Hz and a high cutoff of 40 Hz.
raw.filter(0.1, 40) # Apply a bandpass filter from 0.1 to 40 Hz
Detecting sleep stages
Next, we apply the yasa sleep stage classification algorithm [1]. The only required input is one channel of EEG data. While the algorithm can additionally receive one channel of EOG data, one channel of EMG data, and the age and gender of the participant, these additional inputs are not required. In fact, the EEG channel accounts for almost all of the classifier’s performance [1].
sls = yasa.SleepStaging(raw, eeg_name="Sticky-C1")
y_pred = sls.predict() # Get the predicted sleep stages
hypno = yasa.Hypnogram(y_pred) # Getting a hypnogram for later
hypno_int = yasa.hypno_str_to_int(y_pred)
hypno_up = yasa.hypno_upsample_to_data(hypno=hypno_int, sf_hypno=(1/30), data=data, sf_data=sf)
Inspecting band power
Sleep stages are typically associated with brain activity in specific EEG frequency bands. For instance, N3 sleep is defined by slow waves in the delta band, whereas REM sleep is associated with increased alpha frequency power. We compute the relative power for each sleep stage (Wake, N1, N2, N3, REM) in six common frequency bands (delta, theta, alpha, sigma, beta).
data_filt = raw.get_data() * 1e6
data_c1_uV = data_filt[6]
bandpower_stages = yasa.bandpower(data_c1_uV, sf = 100, win_sec=4, relative=True, hypno=hypno_up, include=(0, 1, 2, 3, 4))
bandpower_avg = bandpower_stages.groupby('Stage')[['Delta', 'Theta', 'Alpha', 'Sigma', 'Beta', 'Gamma']].mean()
bandpower_avg.index = ['Wake', 'N1', 'N2', 'N3', 'REM']
Inspecting the relative band power, we observe increased delta and decreased theta activity in N3 sleep compared to being awake. Further, increased alpha activity is observed in N1 and REM sleep compared to the deeper N2 and N3 sleep stages.
print(bandpower_avg)
Delta Theta Alpha Sigma Beta Gamma
Wake 0.689047 0.252754 0.041008 0.013561 0.003619 0.000011
N1 0.805558 0.156776 0.026298 0.008975 0.002385 0.000009
N2 0.860466 0.122272 0.011644 0.004467 0.001142 0.000010
N3 0.934785 0.058335 0.004424 0.001922 0.000521 0.000014
REM 0.848217 0.115585 0.025746 0.008503 0.001937 0.000010
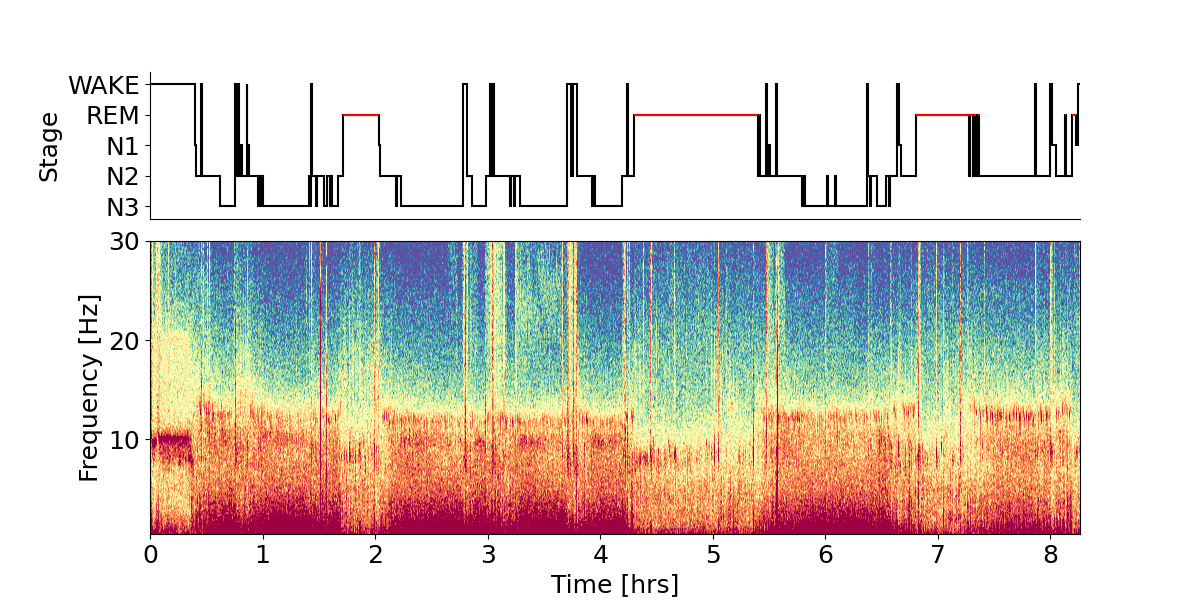
Spectrogram and hypnogram
A hypnogram is a visual representation of the sleep stages over the course of the night. In Figure 1 the hypnogram is combined with a spectrogram displaying the frequency content over time. The hypnogram exhibits a typical pattern of deeper sleep stages in the first half of night and longer REM stages in the second half of the night. The spectrogram reveals power in specific frequency bands that the sleep stages are associated with. Note that usually, clinical sleep recordings are tagged by trained professionals according to the AASM guidelines (American Academy of Sleep Medicine), whereas the hypnogram shown here is based on the classifier’s automatic sleep detection.
fig = yasa.plot_spectrogram(data_c1_uV, sf, hypno=hypno_up, fmax=30, cmap='Spectral_r', trimperc=5)
fig.show()
Slow wave and sleep spindles detection
Slow waves are a hallmark of the N3 sleep stage (also known as slow wave sleep) and, with frequencies between 0.5 and 3 Hz, are part of the delta frequency band. The yasa toolbox provides an slow wave detection algorithm (based on [2, 3]) that allows to inspect individual and averaged slow waves.
Calling the slow wave summary function yields an overview of the timing and properties of the detected slow waves.
sw = yasa.sw_detect(data_c1_uV, sf, hypno=hypno_up)
sw.summary()
Start NegPeak MidCrossing PosPeak End Duration ValNegPeak ValPosPeak PTP Slope Frequency Stage Channel IdxChannel
0 1579.324 1579.600 1579.860 1580.076 1580.400 1.076 -110.009728 70.868551 180.878280 695.685692 0.929368 2 CHAN000 0
1 1833.080 1833.328 1833.560 1833.844 1834.144 1.064 -43.119826 50.537236 93.657062 403.694232 0.939850 2 CHAN000 0
2 1834.144 1834.452 1834.736 1834.896 1835.060 0.916 -64.484388 26.909410 91.393798 321.809148 1.091703 2 CHAN000 0
3 1895.844 1896.224 1896.560 1896.808 1897.060 1.216 -55.624535 42.378489 98.003024 291.675667 0.822368 2 CHAN000 0
4 1935.872 1936.280 1936.700 1936.832 1936.984 1.112 -83.590539 10.574031 94.164569 224.201355 0.899281 2 CHAN000 0
.. ... ... ... ... ... ... ... ... ... ... ... ... ... ...
966 29275.144 29275.496 29275.784 29276.000 29276.756 1.612 -123.916814 68.232216 192.149031 667.184134 0.620347 2 CHAN000 0
967 29276.756 29277.024 29277.292 29277.484 29277.780 1.024 -80.189417 37.322052 117.511469 438.475632 0.976562 2 CHAN000 0
968 29351.656 29351.996 29352.252 29352.468 29352.712 1.056 -80.538877 63.521518 144.060395 562.735919 0.946970 2 CHAN000 0
969 29353.720 29354.168 29354.448 29354.660 29354.980 1.260 -60.494303 36.531033 97.025336 346.519058 0.793651 2 CHAN000 0
970 29444.048 29444.400 29444.704 29444.924 29445.248 1.200 -166.011564 85.047673 251.059236 825.852751 0.833333 2 CHAN000 0
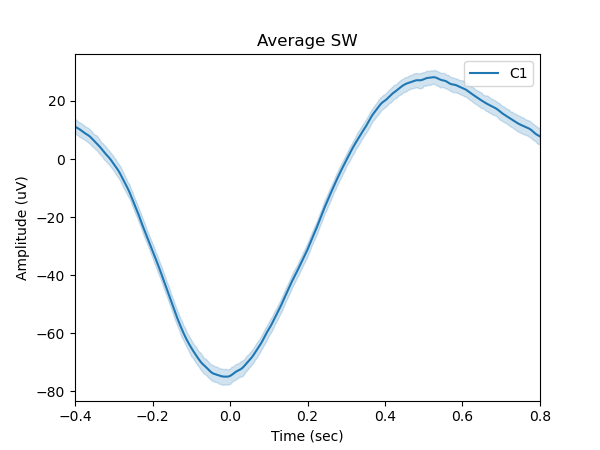
Figure 2 shows the average slow wave, computed from 971 detected slow wave events.
sw.plot_average(time_before=0.4, time_after=0.8, center="NegPeak");
plt.legend(['C1'])
plt.show()
Sleep spindles are a short oscillatory bursts that typically occur during N2 sleep and fall in the frequency range between 11 and 16 Hz. Using the same workflow as before we run the spindle detection algorithm (based on [4]). Calling the sleep spindle summary function yields an overview of the timing and properties of the detected sleep spindles.
sp = yasa.spindles_detect(data_c1_uV, sf)
sp.summary()
Start Peak End Duration Amplitude RMS AbsPower RelPower Frequency Oscillations Symmetry Channel IdxChannel
0 8.720 8.864 9.396 0.676 56.961351 11.090094 2.100221 0.322695 11.315217 8.0 0.211765 CHAN000 0
1 53.476 53.532 54.152 0.676 147.736709 28.034810 2.648341 0.305501 12.936249 8.0 0.082353 CHAN000 0
2 1399.952 1400.424 1400.636 0.684 58.795353 11.988885 1.960096 0.266393 13.047958 8.0 0.686047 CHAN000 0
3 1444.672 1444.860 1445.176 0.504 49.023655 11.592732 2.043340 0.243198 13.838615 7.0 0.370079 CHAN000 0
4 1496.024 1496.072 1496.672 0.648 37.978448 10.030894 1.689857 0.238392 13.444519 8.0 0.073620 CHAN000 0
.. ... ... ... ... ... ... ... ... ... ... ... ... ...
519 29412.916 29413.096 29413.660 0.744 37.617005 8.592611 1.851378 0.323409 12.606909 8.0 0.240642 CHAN000 0
520 29418.196 29419.088 29419.892 1.696 73.779483 14.525022 2.380002 0.519697 12.552197 21.0 0.524706 CHAN000 0
521 29423.800 29425.016 29425.484 1.684 58.250863 11.203151 1.982496 0.389862 12.449155 21.0 0.720379 CHAN000 0
522 29451.216 29452.136 29452.268 1.052 46.337193 10.750559 2.002255 0.438899 12.765855 13.0 0.871212 CHAN000 0
523 29467.156 29467.684 29468.148 0.992 49.976640 9.770195 1.971178 0.429425 12.662866 12.0 0.530120 CHAN000 0
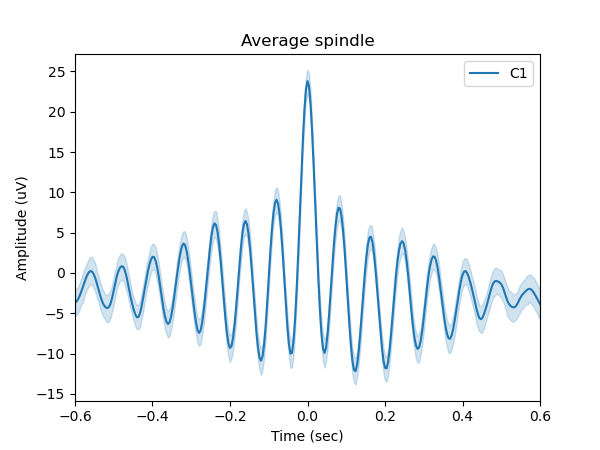
Figure 3 shows the average sleep spindle, computed from 524 detected spindle events.
sp.plot_average(time_before=0.6, time_after=0.6);
plt.legend(['C1'])
plt.show()
Closing words
Whether conducting research in a controlled laboratory environment, studying sleep in an in-home setting, or building a sleep monitoring application, our Mentalab Explore system provides research-grade data quality and the efficiency needed to extract meaningful insights from sleep ExG data. For previous applications of Mentalab Explore in sleep science, we recommend the work by Biller et al. [5] who conduct a longitudinal sleep study and Hainke et al. [6], who investigated gamma oscillations and consciousness during sleep using steady-state visually evoked potentials (SSVEPs) during sleep.
For more information or support, do not hesitate to get in contact at: support@mentalab.com
References
[1] Vallat, R., & Walker, M. P. (2021). An open-source, high-performance tool for automated sleep staging. eLife, 10, e70092. https://doi.org/10.7554/eLife.70092
[2] Carrier, J., Viens, I., Poirier, G., Robillard, R., Lafortune, M., Vandewalle, G., Martin, N., Barakat, M., Paquet, J., & Filipini, D. (2011). Sleep slow wave changes during the middle years of life. European Journal of Neuroscience, 33(4), 758–766. https://doi.org/10.1111/j.1460-9568.2010.07543.x
[3] Massimini, M., Huber, R., Ferrarelli, F., Hill, S., & Tononi, G. (2004). The Sleep Slow Oscillation as a Traveling Wave. Journal of Neuroscience, 24(31), 6862–6870. https://doi.org/10.1523/JNEUROSCI.1318-04.2004
[4] Lacourse, K., Delfrate, J., Beaudry, J., Peppard, P., & Warby, S. C. (2019). A sleep spindle detection algorithm that emulates human expert spindle scoring. Journal of Neuroscience Methods, 316, 3–11. https://doi.org/10.1016/j.jneumeth.2018.08.014
[5] Biller, A. M., Fatima, N., Hamberger, C., Hainke, L., Plankl, V., Nadeem, A., Kramer, A., Hecht, M., & Spitschan, M. (2024). The Ecology of Human Sleep (EcoSleep) Project: Protocol for a longitudinal cohort repeated-measurement-burst study to assess the relationship between sleep determinants and sleep outcomes under real-world conditions across time of year (p. 2024.02.09.24302573). medRxiv. https://doi.org/10.1101/2024.02.09.24302573
[6] Hainke, L., Herberger, S., Taylor, P., & Dowsett, J. (2022). Studying Gamma oscillations and consciousness during sleep using SSVEPs. https://doi.org/10.13140/RG.2.2.35554.81609